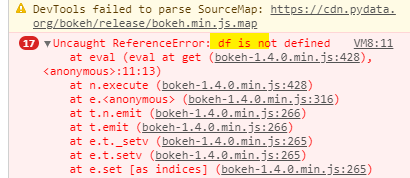
Yes, that helped!
I have been cont. working on the dashboard and in the previous code I did some modelling on the data. I have defined the model below. Do I need to do this in js to get the results for A and n for the selected data? Tried to find an example of it in bokeh, but can only find related to bokeh serve.
To have a standalone html, I suspect I need to add the model to each customjs below?
I had something like this previously (rest of code is below):
time = np.arange(start = 1, stop = int(round(df.Average_time_hours.max())) + 500, step = 10)
y = releasePowerFunction(time, df['A-value'].unique(), df['n-value'].unique())
p.line(time, y, color = 'black')
### rest of code:
df = pd.DataFrame({'MC_Temperature': [20, 40, 80, 10, 10, 20.5, 50.0, 50, 10, 10],
'Matrix_Name': ['RM-006', 'LAT-200/LTA-202 80/20', 'LAT-220', 'LAT-200', 'LAT-200/LTA-202 80/20', 'LAT-220', 'LAT-200', 'LAT-200/LTA-202 80/20', 'LAT-220', 'LAT-220'],
'Tracer_Content': [10.0, 10.0, 1.0, 10.0, 2.3, 1, 1, 2, 1, 2],
'Average_time_hours': [10, 2, 1, 8, 10, 2, 1, 8, 10, 2],
'Coefficiant of Variance [%]': [10, 2, 1, 8, 10, 2, 1, 8, 10, 2],
'Release [mg/(cm2*d)]': np.random.randint(low=1, high=100, size=10)})
df.MC_Temperature = df.MC_Temperature.astype(str)
df.Tracer_Content = df.Tracer_Content.astype(str)
source = ColumnDataSource(df)
powerModel = Model(releasePowerFunctionLog)
pars = powerModel.make_params(A = 1, n = 1)
def model(df):
df = df[df["Release [mg/(cm2*d)]"] > 0]
R = np.log(df[['Release [mg/(cm2*d)]']].values)
t = np.log(df[['Average_time_hours']].values)
fits = powerModel.fit(R, pars, method = 'leastsq', t = t)
df['A-value'] = np.exp(fits.params['A'])
df['n-value'] = np.float(fits.params['n'])
return df#np.exp(fits.params['A']), np.float(fits.params['n'])
def add_multiselect_filter_callback(value, columnName):
value_filter = BooleanFilter([True if x == value else False for x in source.data[columnName]])
selectoroptions = [str(x) for x in sorted(np.unique(source.data[columnName]))]
multi_select = MultiSelect(title= columnName, value = [value],
options=selectoroptions)
return value_filter, multi_select
Temperature = '60.0'
temp_filter, temp_multi_select = add_multiselect_filter_callback(Temperature, 'MC_Temperature')
temp_multi_select.js_on_change('value', CustomJS(args = dict(f=temp_filter, source=source, columnName = 'MC_Temperature'),
code="""\
const val = cb_obj.value;
f.booleans = Array.from(source.data[columnName]).map(d => val.includes(d != null && d.toString()));
source.change.emit();
"""))
tracerContent = '10.0'
tc_filter, tc_multi_select = add_multiselect_filter_callback(tracerContent, 'Tracer_Content')
tc_multi_select.js_on_change('value', CustomJS(args = dict(f=tc_filter, source=source, columnName = 'Tracer_Content'),
code="""\
const val = cb_obj.value;
f.booleans = Array.from(source.data[columnName]).map(d => val.includes(d != null && d.toString()));
source.change.emit();
"""))
matrixName = 'RM-006'
mn_filter, mn_multi_select = add_multiselect_filter_callback(matrixName, 'Matrix_Name')
mn_multi_select.js_on_change('value', CustomJS(args = dict(f=mn_filter, source=source, columnName = 'Matrix_Name'),
code="""\
const val = cb_obj.value;
f.booleans = Array.from(source.data[columnName]).map(d => val.includes(d != null && d.toString()));
source.change.emit();
"""))
p = figure(tools = ["box_zoom", "hover", "reset", "save", "crosshair", "pan"],
plot_width = 1600,
plot_height = 600,
y_axis_type = "log",
y_axis_label = 'Release [mg/(cm2*d)]')
p2 = figure(plot_width=1600, plot_height=200, background_fill_color="#fafafa", x_range=p.x_range,
x_axis_label = 'Time [h]',
y_axis_label = 'Coefficiant of Variance [%]')
p.circle(x='Average_time_hours', y='Release [mg/(cm2*d)]', source=source, view=CDSView(source=source, filters=[temp_filter, tc_filter, mn_filter]), color='black')
p2.scatter(x = "Average_time_hours", y = 'Coefficiant of Variance [%]', color = 'green', marker = 'hex', source = source, view=CDSView(source=source, filters=[temp_filter, tc_filter, mn_filter]))
p2.line(x = "Average_time_hours", y = 'Coefficiant of Variance [%]', color = 'green', line_dash = 'dashed', source = source, view=CDSView(source=source, filters=[temp_filter, tc_filter, mn_filter]))
columns = [
TableColumn(field="MC_Sample_ID", title="LTO", width=150),
TableColumn(field="MC_ID", title="MC ID", width=100),
TableColumn(field="MC_Temperature", title="Temperature [°C]", editor=IntEditor()),
TableColumn(field="Matrix_Name", title="Matrix Name", formatter=StringFormatter(font_style="bold"),width=450),
TableColumn(field="Tracer_Content", title="Tracer Content", formatter=StringFormatter(font_style="bold"),width=450),
TableColumn(field="RESMAN_Tracer_Code", title="Tracers", formatter=StringFormatter(font_style="bold"), width=150),
TableColumn(field="Time_Input", title="Time Input", editor=IntEditor()),
TableColumn(field="Molecular_Weight", title="Molecular Weight", editor=IntEditor()),
TableColumn(field="Average_time_hours", title="Average time hours", editor=IntEditor()),
TableColumn(field="A-value", title="A-value", editor=IntEditor()),
TableColumn(field="n-value", title="n-value",editor=IntEditor()),
TableColumn(field="Release [mg/(cm2*d)]", title="Release [mg/(cm2*d)]", editor=IntEditor()),
TableColumn(field="Coefficiant of Variance [%]", title="Coefficiant of Variance [%] ", editor=IntEditor()),
]
data_table = DataTable(source = source, columns = columns, view=CDSView(source=source, filters=[temp_filter, tc_filter, mn_filter]), width=1600)
output_file('tester.html')
layout = row(column(p, p2, data_table), column(temp_multi_select, tc_multi_select, mn_multi_select, Spacer(width=400, height=500)))
show(layout)



 Before you fix it, open the JavaScript console in your browser and try to select something. You should see an error there.
Before you fix it, open the JavaScript console in your browser and try to select something. You should see an error there.