Hello,
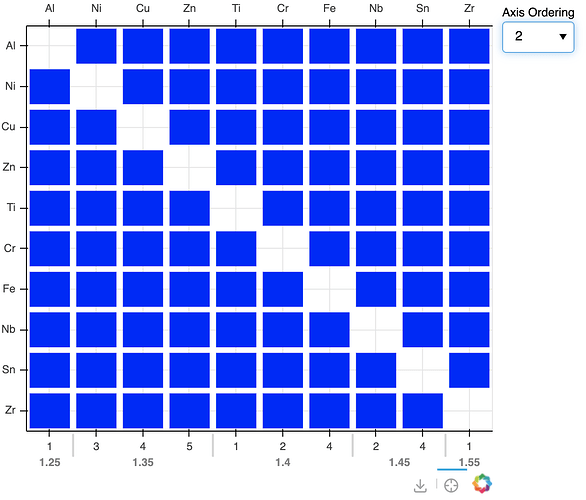
I am building a figure for my research project that displays a matrix of 70 elements on both the x and y axes of the figure to display information elemental interactions at the intersections.
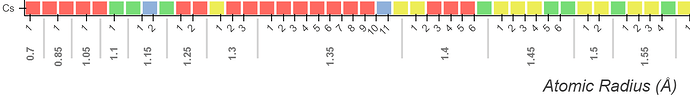
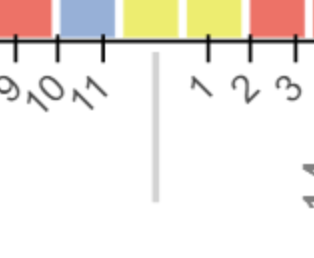
I have previously built functionality to sort the axes based on elemental properties (i.e. ascending atomic radius) via a CustomJS callback. I recently decided that I should also display the values that the elements are being sorted on, so I created an extra x range for this information and added it to the bottom of the figure using add_layout(). The elements are arranged on a CategoricalAxis, so I decided to use a Categorical Axis for the bottom axis that displays elemental properties. This is advantageous because I want to create suborderings, and sometimes the data is non-numerical if it doesn’t exist.
I am now running into an issue where I want to have evenly spaced ticks, but when I update the figure.x_range.factors in the JS callback to have a subordering, the ticks are no longer spaced linearly. I have tried switching to BaseTicker or FixedTicker by passing in an instance in the arguments of the CategoricalAxis constructor, but the figure crashes in both instances. Next, I tried creating a copy of the top x_range ticker using .clone() in the JS portion so that it overwrites any automatic scaling after the factors are updated, but that had no effect on the tick spacing.
Please let me know if there is any way to fix this!
Here is a simplified version of the code:
from bokeh.io import show
from bokeh.layouts import row
from bokeh.models import ColumnDataSource, CategoricalAxis, Select, CustomJS, FactorRange
from bokeh.plotting import figure
tbt = ['Ti', 'Fe', 'Al', 'Ni', 'Cr', 'Cu', 'Zr', 'Nb', 'Zn', 'Sn']
xnames = []
ynames = []
for i in tbt:
for j in tbt:
if i == j:
continue
xnames.append(i)
ynames.append(j)
dsource = ColumnDataSource(data=dict(
xname=xnames,
yname=ynames
))
p = figure(x_axis_location="above", tools=["save", "crosshair"],
x_range=tbt,
y_range=list(reversed(tbt)),
toolbar_location="below", width=500, height=500)
p.rect(x='xname', y='yname', width=0.86, height=0.86, color="blue", line_width=0.06, source=dsource)
axis_orderings = {"1": [['Al', '13'], ['Ti', '22'], ['Cr', '24'], ['Fe', '26'], ['Ni', '28'], ['Cu', '29'],
['Zn', '30'], ['Zr', '40'], ['Nb', '41'], ['Sn', '50']],
"2": [['Al', ['1.25', '1']], ['Ni', ['1.35', '3']], ['Cu', ['1.35', '4']],
['Zn', ['1.35', '5']], ['Ti', ['1.4', '1']], ['Cr', ['1.4', '2']], ['Fe', ['1.4', '4']],
['Nb', ['1.45', '2']], ['Sn', ['1.45', '4']], ['Zr', ['1.55', '1']]]
}
p.extra_x_ranges['bottom'] = FactorRange(factors=[data[1] for data in axis_orderings["1"]])
p.add_layout(CategoricalAxis(x_range_name='bottom'), 'below')
axis_select = Select(title="Axis Ordering", value="1", options=list(axis_orderings.keys()))
axis_callback = CustomJS(args=dict(select=axis_select, p=p, orderings=axis_orderings), code="""
var ordering_name = select.value;
var element_symbol_order = orderings[ordering_name].map(sub_arr => sub_arr[0]);
var element_property_labels = orderings[ordering_name].map(sub_arr => sub_arr.slice(1)).flat();
console.log(element_symbol_order);
console.log(element_property_labels);
p.x_range.factors = element_symbol_order.slice();
p.y_range.factors = element_symbol_order.slice().reverse();
p.extra_x_ranges['bottom'].factors = element_property_labels.slice();
""")
axis_select.js_on_change('value', axis_callback)
r = row(p, axis_select)
show(r)
Elemental symbol axis (bottom):