Hi,
I have a squared image which is essentially a 2d numpy array. I want to highlight different parts of the image according to its value in each pixel.
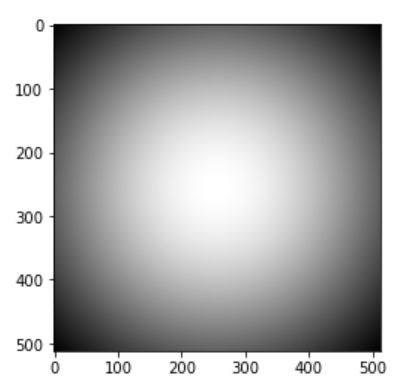
This is the an example image: 2d guassian
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
x, y = np.meshgrid(np.linspace(-1,1,512), np.linspace(-1,1,512))
d = np.sqrt(x*x+y*y)
sigma, mu = 1.0, 0.0
g = np.exp(-( (d-mu)**2 / ( 2.0 * sigma**2 ) ) )
plt.imshow(g, 'gray')
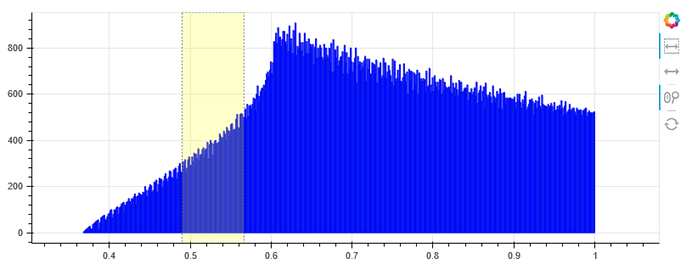
This is its intensity histogram
import numpy as np
from bokeh.io import show, output_notebook
from bokeh.layouts import column
from bokeh.models import ColumnDataSource, RangeTool
from bokeh.plotting import figure
output_notebook()
sample_img_nonan = g[~np.isnan(g)]
hist, edges = np.histogram(sample_img_nonan, bins=500)
p = figure(plot_height=300, plot_width=800,
tools='xpan, xwheel_zoom, reset',
active_scroll='xwheel_zoom',
active_drag='xpan')
p.quad(top=hist, bottom=0, left=edges[:-1], right=edges[1:], line_color="blue")
range_tool = RangeTool(x_range=Range1d(np.mean(sample_img_nonan)-np.std(sample_img_nonan),
np.mean(sample_img_nonan)+np.std(sample_img_nonan)))
range_tool.overlay.fill_color = "yellow"
range_tool.overlay.fill_alpha = 0.2
p.add_tools(range_tool)
p.toolbar.active_multi = range_tool
show(p)
The yellow rectangular can be adjusted in size and slided along the x direction, and I want to use it to specify the intensity range which will be highlighted on the origin image. Like below:
mask = np.ma.masked_outside(g, 0.75, 0.83 )
fig, ax = plt.subplots(figsize=(10,10))
ax.imshow(g, 'gray')
ax.imshow(mask,'gist_rainbow', interpolation='none', alpha=0.7)
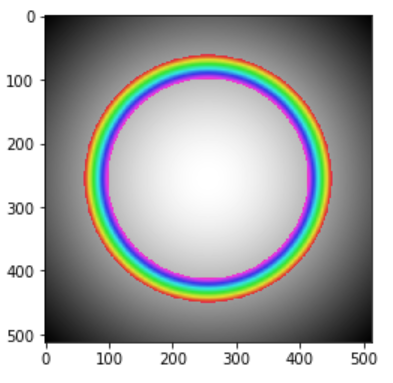
Now I have no idea how to link the histogram to the image, I have looked into here and here. But I’m not sure if that is the right direction. Please give me some hint so I can go into the right direction for this issue. Thanks in advanced!