Hello,
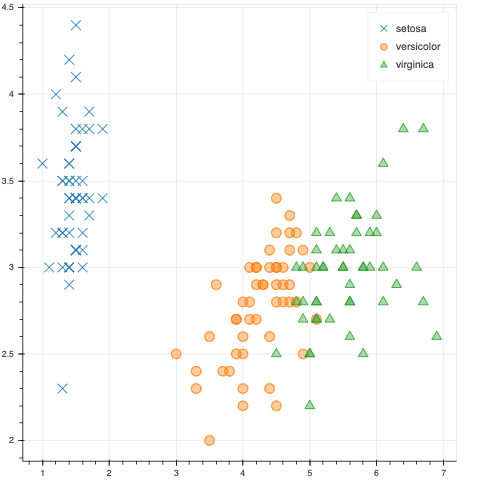
I’m using scatter to make an interactive plot. Since hiding individual glyphs doesn’t seem to work with this type of plot, I’ve implemented some checkboxes to include/remove certain subsets of the data. I’m using legend_group to create the legend.
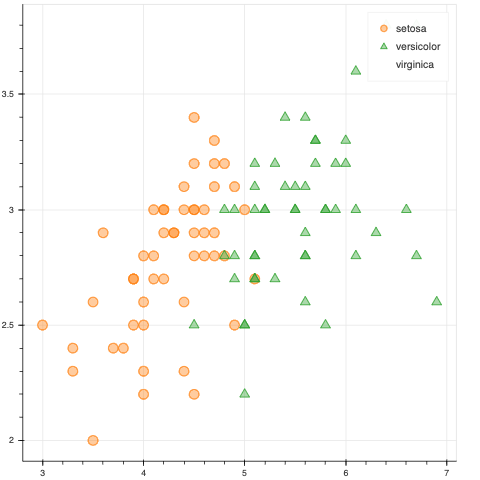
The problem is, when I subset the data, the legend labels stay put, but the glyph of the removed group is removed and the remaining labels get shifted. Is this the expected behavior?
I have modified the code from this example to show what is happening.
If there is a better way to get this done, please let me know!
Thank you!
from bokeh.models import Legend, LegendItem, ColumnDataSource
from bokeh.palettes import Category10_3
from bokeh.plotting import figure, show
from bokeh.sampledata.iris import flowers
from bokeh.transform import factor_cmap, factor_mark
from bokeh.io import curdoc
from bokeh.models.widgets import CheckboxGroup
from bokeh.layouts import column
flowers_ss = flowers.copy()
flower_source = ColumnDataSource(flowers)
def update_data(attr,old,new):
s_match = '|'.join([SPECIES[x] for x in new])
flowers_ss = flowers[flowers['species'].str.contains(s_match)]
flower_source.data = dict(
sepal_length=flowers_ss['sepal_length'],
sepal_width=flowers_ss['sepal_width'],
petal_length=flowers_ss['petal_length'],
petal_width=flowers_ss['petal_width'],
species=flowers_ss['species'])
SPECIES = ['setosa', 'versicolor', 'virginica']
MARKERS = ['x', 'circle', 'triangle']
p = figure()
# plot the actual data using factor and color mappers (using the same
# column `species` here but you can use two different columns if you want)
r = p.scatter("petal_length", "sepal_width",
source=flower_source, fill_alpha=0.4, size=12,
legend_group='species',
marker=factor_mark('species', MARKERS, SPECIES),
color=factor_cmap('species', Category10_3, SPECIES))
species_select = CheckboxGroup(
labels=SPECIES,
active=list(range(len(SPECIES))))
species_select.on_change('active', update_data)
curdoc().add_root(column(p, species_select))